Protein Structure Determination
Solutions NMR is the only technique to get a three-dimensional structure for a protein or protein ligand complex in its native functional state. In addition to providing insights about their three-dimensional structures and functions, it can also be employed to probe the protein or enzyme active sites and folding changes associated with enzyme activation.
The zinc finger motif was deduced from the pattern of sequence conservation in SAP30 orthologs. The motif and the fold is novel in that it shares little or no similarity with other well-characterized zinc finger motifs. The functional role of the zinc finger was deduced via informatics and NMR approaches. A distinctive feature of the domain is the presence of a highly conserved surface (shown in green, middle panel) that coincides with a highly basic region. Residues in the highly conserved, basic region exhibit strong perturbations in NMR titration experiments (shown in deep yellow and magenta in the right panel) when presented with diverse oligonucleotide substrates, suggesting a role for the zinc finger motif in oligonucleotide binding.
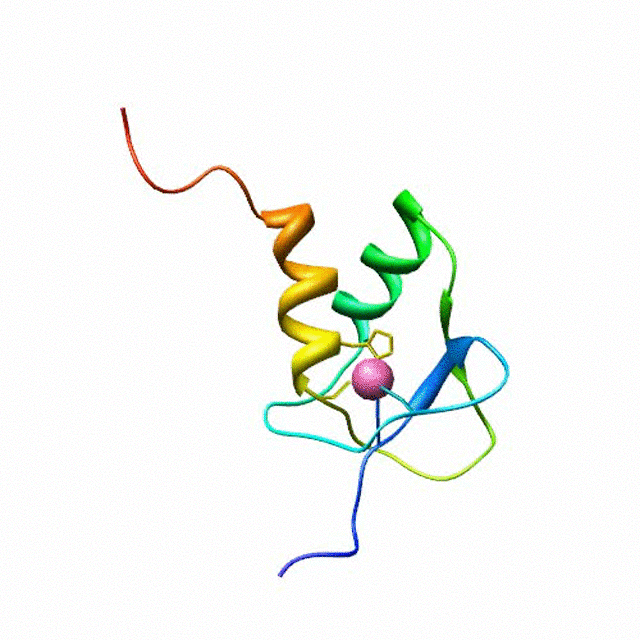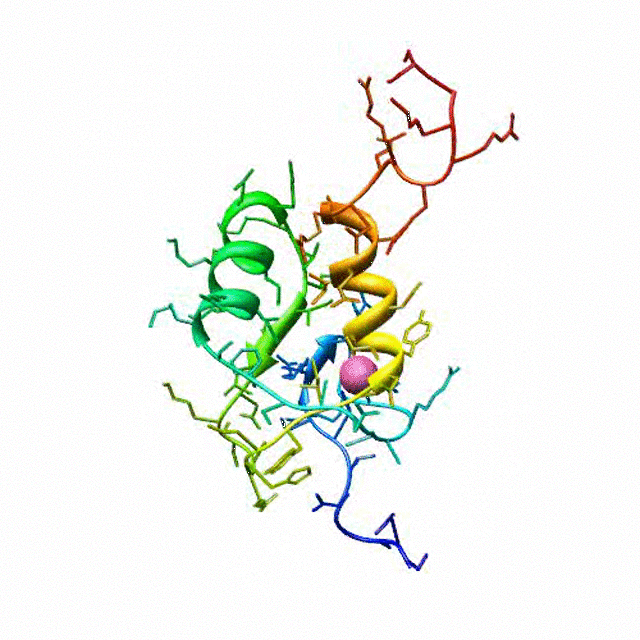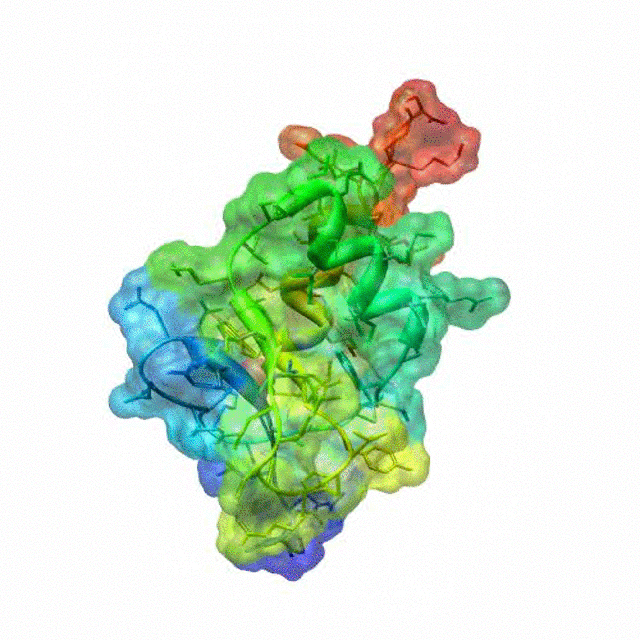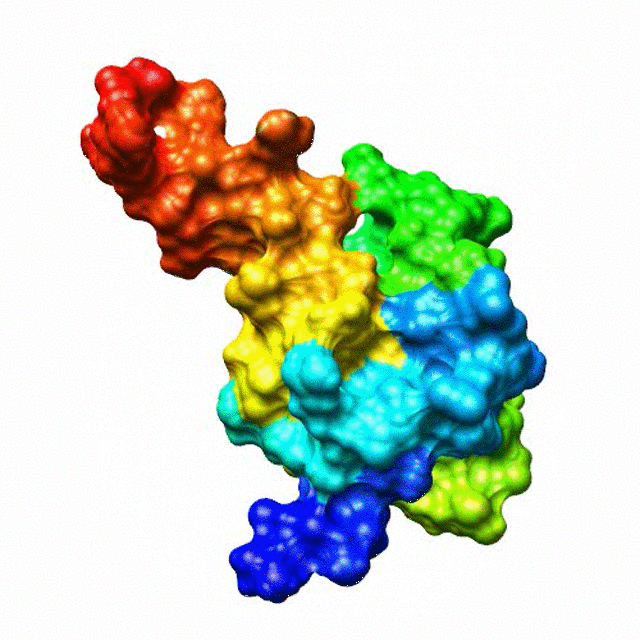 A solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex. Courtesy of the Prof. Ishwar Radhakrishnan Lab (
Source
).
A solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex. Courtesy of the Prof. Ishwar Radhakrishnan Lab (
Source
).
Usually the proteins are expressed with E.Coli in either rich medium or isotopically enriched medium (13C glucose and 15N NH4Cl) and purified by FPLC or HPLC. The NMR sample is normally buffered at pH below 7. The volume is 500 uL. It should contain 10% D2O for locking purpose.
If you want to start a structure biology project, please click here.
Start a project