Biomolecular NMR
NMR structure biology
With modern digital NMR equipment, rapid protein structure determination with solution NMR is a reality for proteins with molecular weight below 25 kDa. It is straight forward to get a three-dimensional solution NMR structure or a protein or protein ligand complex in its native functional state. In addition to providing insights about their three-dimensional structures and functions, it can also be employed to probe the protein or enzyme active sites and folding changes associated with enzyme activation. To complete NMR protein experiments, your protein needs to be soluble the tens of micromolar (uM) up to millimolar (mM) range. Stable isotope labeling is required for higher molecular weight proteins. IMSERC can help with generation of these compound in collaboration with the
Recombinant Protein Production Core.
More details on how to assess the stability and folding of proteins
.
Case Studies
Following are two animations demonstrate the structure and dynamics of proteins using solution NMR.
Animation 1: Zinc Finger
The zinc finger motif was deduced from the pattern of sequence conservation in SAP30 orthologs. The motif and the fold is novel in that it shares little or no similarity with other well-characterized zinc finger motifs. The functional role of the zinc finger was deduced via informatics and NMR approaches. A distinctive feature of the domain is the presence of a highly conserved surface (shown in green, middle panel) that coincides with a highly basic region. Residues in the highly conserved, basic region exhibit strong perturbations in NMR titration experiments (shown in deep yellow and magenta in the right panel) when presented with diverse oligonucleotide substrates, suggesting a role for the zinc finger motif in oligonucleotide binding.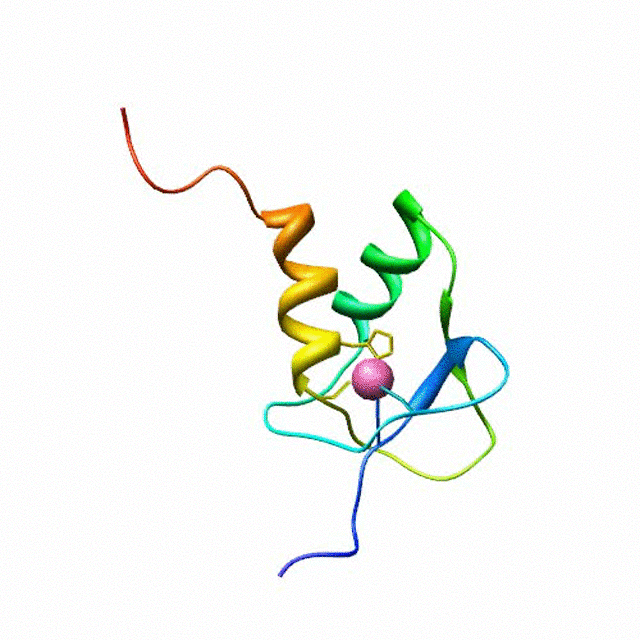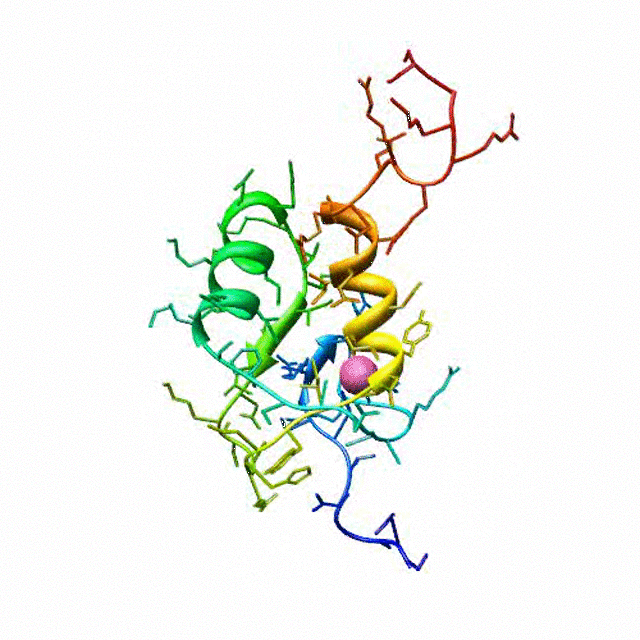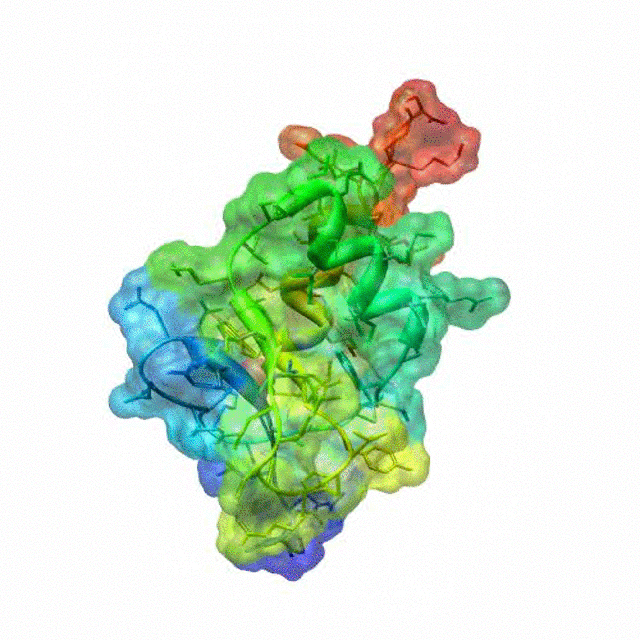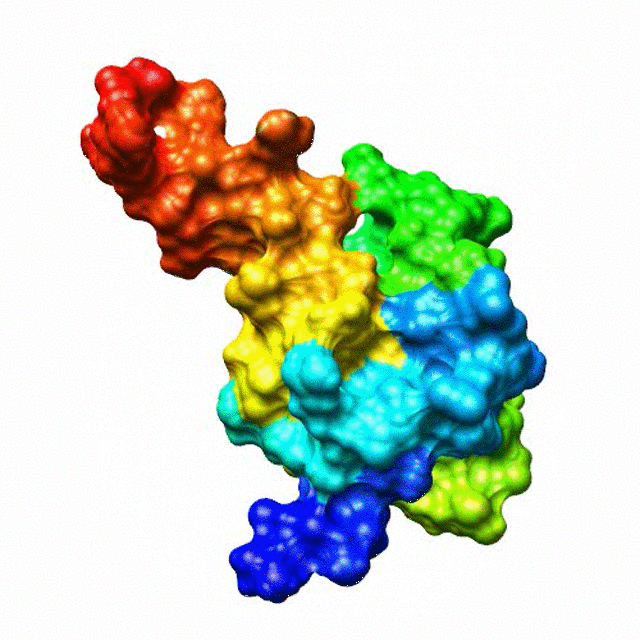 A solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex. Courtesy of the Prof. Ishwar Radhakrishnan Lab (
Source
).
A solution structure of a novel zinc finger motif in the SAP30 polypeptide of the Sin3 corepressor complex. Courtesy of the Prof. Ishwar Radhakrishnan Lab (
Source
).
Animation 2: The contortion of El TAR
This animation is derived based on domain-elongated NMR RDC data. Shown in gray are the various conformations TAR RNA takes when bound to different small molecules, including ligands designed to combat HIV. The video shows how the RNA molecule passes through various critical conformations in the process of bending and twisting.
IMSERC Staff are available to assist with sample preparation protocols and choice of techniques for protein structure elucidation. IMSERC collaborates with NU's Center for Structural Biology .
