|
|
From a proton (1H) spectrum information about chemical shifts, multiplet structures, homonuclear (and heteronuclear) coupling constants and integration of all protons present in the sample can be obtained. In addition, line widths are related to relaxation T2 mechanisms. Alternatively, other related experiments can give extra information as, for instance, variable temperature experiments, the use of relaxing and/or complexing reagents ...REQUIREMENTS
Easy implementation on AVANCE spectrometerVERSIONS
The basic single-pulse sequence to record a standard proton spectrum consists of the following steps:EXPERIMENTAL DETAILSRelaxation period (d1) to achieve a pre-equilibrium state. Read pulse (p1) to create transverse magnetization. Acquisition during the t2 (acq) period. When high-abundant X heteronuclei are present, broadband or selective X-decoupling can be applied from a second channel. When high-abundant X heteronuclei are present, broadband or selective X-decoupling can be applied during d1 and/or the acquisition periods from a second channel.
The proton spectrum is usually acquired, processed and plotted in a fully automated way. Minor changes from a predefined parameter set are required. For routine applications, a 30º-45º pulse with a short relaxation delay (1 second) is usually applied. In some demanding applications, good accuracy is advisable to obtain reliable integration values. In this case, a proton spectrum is recorded with a 90º proton pulse and a long relaxation period (5*T1(1H)) is used to avoid partial signal saturation.
In samples containing high-abundant heteronuclei as 31P, 19F ..., an X-decoupled
1H spectrum can be helpful:
The proton spectrum displays resonance intensities vs frequency. The signals are referenced to the TMS signal (0 ppm) or to the internal deuterated solvent. Usually, chemical shift (resonance position) and coupling constants (lines separation within a multiplet) are extracted.RELATED TOPICS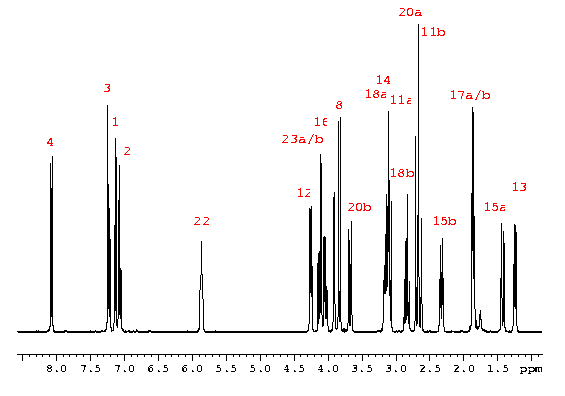
See More Spectra
Full assignment of proton spectra is usually accomplished by the concerted use with other NMR methods. As a general strategy:
- Through-bond correlation with other protons via J(HH) could be established from homodecoupling, 2D COSY or 2D TOCSY spectra.
- Through-space correlations with other protons via NOE is achieved from NOE-type (see NOESY) or ROE-type (see ROESY) experiments.
- Heteronuclear correlation with the directly attached X nuclei via 1J(XH) is achieved from HETCOR and inverse-detected correlation (see HMQC and HSQC experiments.
- Long-Range correlation through two and three bonds with X nuclei via nJ(CH) is achieved from COLOC and HMBC experiments.