|
|
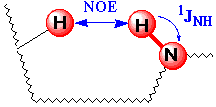
The 3D NOESY-HSQC experiment is specifically designed to obtain X-edited NOESY spectra of X-labeled biomolecules and protein-ligand complexes from which homonuclear 1H-1H NOEs can be clearly assigned even in overcrowded regions (the mechanism involves 1H-1H NOE step followed by an heteronuclear transfer via 1J(XH)). Similar results can be obtained from an analog 3D NOESY-HMQC experiment.REQUIREMENTS
Implementation on AVANCE spectrometers is feasible. Improved versions using pulsed field gradients (PFGs) are also available and, therefore, gradient technology is required. The 1H-13C NOESY-HSQC experiment is usually applied on 13C-labeled proteins in D20. and the 1H-15N NOESY-HSQC experiment is usually applied on 15N-labeled molecules in H20.VERSIONS
The original NOESY-HSQC pulse sequence basically consists of an homonuclear 2D NOESY pulse sequence followed by a 1H-X HSQC pulse sequence. However, the HSQC block could be replaced with other schemes: decoupled HSQC, CT-HSQC, PEP-HSQC, HMQC ...(see, for instance, the 3D NOESY-HMQC experiment).EXPERIMENTAL DETAILSSeveral improved versions have been proposed incorporating the following modifications:
- 3D NOESY-refocused HSQC experiment ( 95JB433 ).
- Use of PFGs ( 93JMRB317-102 ) combined with solvent suppression using WATERGATE ( 93JMRA241-102 and 95JMRB238-109 ) and PEP methodology ( 95JMRB86-106 ).
- Improved sensitivity using a 45 degree water flip back pulse ( 99JMR451-140 ).
- Incorporation of theTROSY approach ( 99JB77 and 99PNAS9607 ). A 2D [1H-13C]-[1H-13C] NOESY-TROSY experiment for simultaneous detection of backbone NH, aromatic CH and side-chain NH2 correlations has been reported ( 00JB195 )
- A w1-filtered version has been proposed that uses an double-tuned X-filter element as a initial period ( 96JB492-8 , 97JACS6711 ).
- Simultaneous evolution of 13C and 15N chemical shift in the t2 period ( 94JMRB197-103 , 94BIO14858 and , 03JB193-27).
- A program called SPIRIT has been developed to simulate 3D NOESY-HSQC experiment ( 98JB17 ).
- Three-way decomposition on 15N-NOESY-HSQC using the software MUNIN ( 02JB191-24 ).
- Increased resolution from a 13C-edited NOESY-HSQC experiment with split carbon evolution ( 98JMR191-132 ).
- Related double- and triple-resonance 3D 13C/15N edited NOESY-HSQC ( 94BIO14858 , 95JMRB294-108 , and 95JMRB238-109 ) and 4D 13C-15N-edited NOESY-HSQC experiments have also been proposed ( 90SCI411 ) to observe NOEs between protons with degenerate chemical shifts.
- The initial NOESY block can be replaced by other homonuclear mixing processes as described in the 3D TOCSY-NOESY-HSQC and NOESY-TOCSY-HSQC experiments ( 92JMR408-96 ).
- A simultaneous 3D NOESY-13C/15N-HSQC has been proposed to determine intra- and intermolecular NOESY in protein ligand complexes ( 01JB107-21 )
The 3D NOESY-HSQC experiment can be recorded in automation mode. More details on practical implementation of the 3D NOESY-HSQC experiment on AVANCE spectrometers can be found in the corresponding Tutorial 3D NOESY-HSQC experimentSPECTRA
The NOESY-HSQC experiment affords a 3D spectrum in which 1H, X and 1H chemical shifts are displayed in three independent dimensions. Thus, the F1(1H)-F3(1H) plane of a 1H-15N NOESY-HSQC spectrum will show a typical 2D F1(1H)-F2(NH) NOESY map. On the other hand, the F1(1H)-F3(1H) plane of a 1H-13C NOESY-HSQC spectrum will show a typical 2D F1(1H)-F2(1H aliphatic) NOESY map while the F1(1H)-F2(13C) plane will correspond to a conventional 2D 1H-13C HSQC spectrum.RELATED TOPICS
General Reading: 96ENC4733Simulation of 15N or 13C edited 3D NOESY-HSQC experiment is integrated in the program AURELIA( 99JMR39-137 )